Agenda:
- Overview of graphs
Terminology¶
A graph $G$ consists of
- a set of $n$ vertices/nodes $V$
- a set of $m$ edges $E \subseteq V \times V$
A graph is way to represent objects and their relations
- Node: represents an object
- Edge: represents a relation between two nodes.
- Neighbor: Two nodes are neighbors if they are connected by an edge.
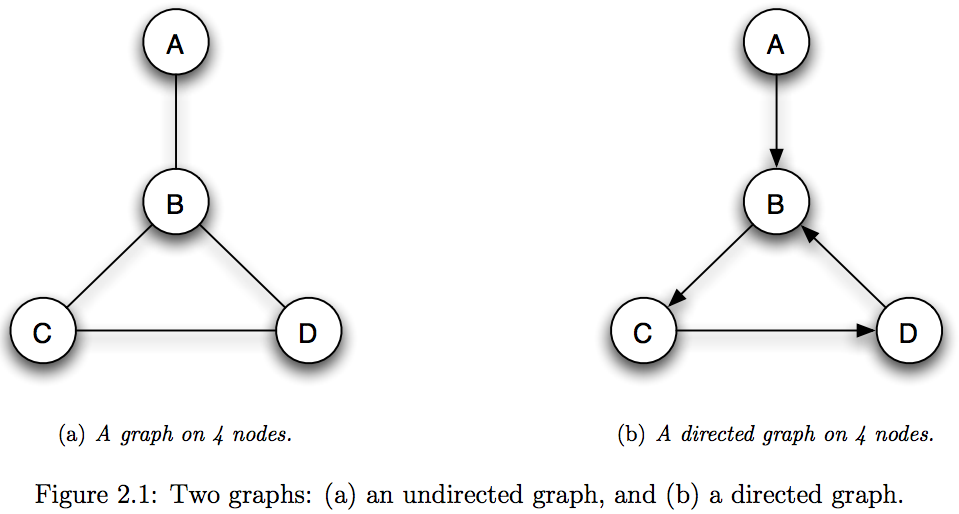
- Directed Graph: Represents asymmetric (one-way) relationships
- Undirected Graph: Represents symmetric relationships
Graphs queries¶
What are some things you might want to know about a graph? E.g., consider social networks, transport network, etc.
Who is friends with Billie Eilish?
find neighbors of a node:
$ N(v) =\{v_i ~ \mid (v, v_i) \in E ~~ \hbox{or} ~~(v_i, v) \in E\}$
How popular is Billie Eilish?
degree: number of neighbors of a node:
$ |N(v)|$
- Do I know someone who is friends with Billie Eilish?
- path: a sequence of nodes in which each consecutive pair are neighbors
- reachability: $a$ is reachable from $b$ if there is a path between them
- Am I in the same clique as Billie Eilish?
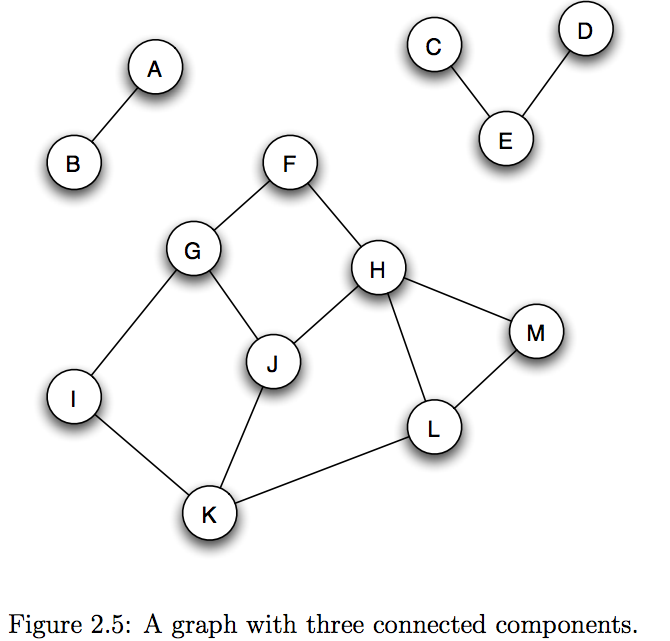
- a graph is connected if there is a path between each pair of nodes
- connected component: A maximal subset of nodes such that each pair of nodes is connected
What data structures can we use to represent a graph?¶
Adjacency matrix¶

$ \begin{bmatrix} & A & B & C & D\\ A & 0 & 1 & 0 & 0\\ B & 1 & 0 & 1 & 1\\ C & 0 & 1 & 0 & 1\\ D & 0 & 1 & 1 & 0\\ \end{bmatrix} \begin{bmatrix} & A & B & C & D\\ A & 0 & 1 & 0 & 0\\ B & 0 & 0 & 1 & 0\\ C & 0 & 0 & 0 & 1\\ D & 0 & 1 & 0 & 0\\ \end{bmatrix} $

import numpy as np
from tabulate import tabulate
labels = ['A', 'B', 'C', 'D']
nodes = [0,1,2,3]
edges = [(0,1), (1,2), (2, 3), (3, 1)]
def make_graph(nodes, edges, directed=False):
graph = np.zeros((len(nodes), len(nodes)), dtype=int)
for e in edges:
graph[e[0], e[1]] = 1
if not directed: # add reverse direction
graph[e[1], e[0]] = 1
return graph
graph = make_graph(nodes, edges, directed=False)
print('undirected')
print(tabulate(graph, labels))
undirected A B C D --- --- --- --- 0 1 0 0 1 0 1 1 0 1 0 1 0 1 1 0
digraph = make_graph(nodes, edges, directed=True)
print('\ndirected')
print(tabulate(digraph, labels))
directed A B C D --- --- --- --- 0 1 0 0 0 0 1 0 0 0 0 1 0 1 0 0
def neighbors_adjacency(graph, node, labels):
result = []
i = labels.index(node)
print(graph[i])
for index, val in enumerate(graph[i]):
if val == 1:
result.append(labels[index])
return result
neighbors_adjacency(graph, 'B', labels)
[1 0 1 1]
['A', 'C', 'D']
Runtime to access neighbors of a node?
$O(|V|)$
Why is this space inefficient?¶
If there are $|V|$ nodes, what is the maximum number of edges?
....but, if a graph is dense, then there's not really any value in representing the data as a graph.
Luckily, most real-world graphs are extremely sparse.
- E.g., you are probably not friends with over 1,000 people.
it's a small world after all...¶
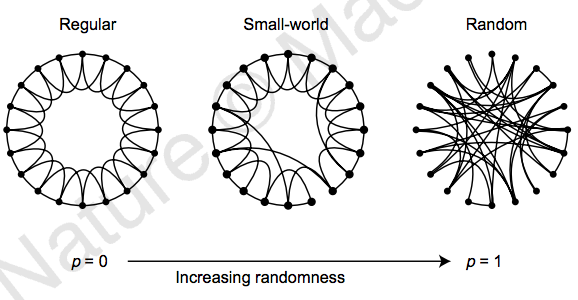
Source: Collective dynamics of 'small-world' networks. Duncan J. Watts & Steven H. Strogatz
Edge Sets¶
We could simply store the graph as a set of edges.
edges = set([('A', 'B'),
('B', 'C'),
('C', 'D'),
('D', 'B')])
edges
{('A', 'B'), ('B', 'C'), ('C', 'D'), ('D', 'B')}
What's the space requirement?
$O(|E|)$
How can we access the neighbors of a node?
def neighbors_set(edges, node):
# assuming an undirected graph
result = []
for e in edges:
if e[0] == node:
result.append(e[1])
elif e[1] == node:
result.append(e[0])
return result
neighbors_set(edges, 'B')
['A', 'D', 'C']
What is the work/span of accessing neighbors using an edge set?
Work: $O(|E|)$
Span: $O(\lg |E|)$ (using filter)
Can we do better?
Map of Neighbors¶
We can use a hashmap (dict) to store the neighbors of each node.
graph = {
'A': {'B'},
'B': {'A', 'C', 'D'},
'C': {'B', 'D'},
'D': {'B', 'C'}
}
graph
{'A': {'B'}, 'B': {'A', 'C', 'D'}, 'C': {'B', 'D'}, 'D': {'B', 'C'}}
What is work of accessing neighbors?
def neighbors_map(graph, node):
return graph[node]
neighbors_map(graph, 'B')
{'A', 'B', 'D'}
Constant time to lookup the neighbors of a node.
Graph Search¶
One of the fundamental operations over graphs
- Start at a source node s
- Visit all reachable nodes
- $t$ is reachable from $s$ if there is a path between them
- For efficiency, visit each node at most once.
Can be used to solve a number of problem:
- Is the graph connected?
- Is node $t$ reachable from node $s$
- Shortest path from $s$ to $t$
Generic Graph Search¶
Consider the task of crawling every web page reachable from a starting page $s$.
How would you do this?
At any point in the search, a vertex can be in one of three sets:
- visited: the set of vertices already visited
- frontier: the unvisited neighbors of the visited vertices
- unseen: everything else
We can then describe a generic search algorithm as follows:
while vertices remain to be visited:
- visit some unvisited nodes in the frontier
- update the three sets
We'll look at two common search algorithms, breadth-first search and depth-first search.
Vertex Hopping¶
Just to get an idea of the problem, we'll start with an inefficient way of searching called vertex hopping
until all nodes visited:
pick a node
visit all its neighborsdef hop(graph):
for v in graph:
print('searching with %s' % v)
for neighbor in graph[v]:
print('...found %s' % neighbor)
hop(graph)
searching with A ...found B searching with B ...found A ...found C ...found D searching with C ...found B ...found D searching with D ...found B ...found C
This of course ignores the idea of paths in a graph.
However, we could still build on this to solve the reachability problem. How? We'll address this problem in recitation.